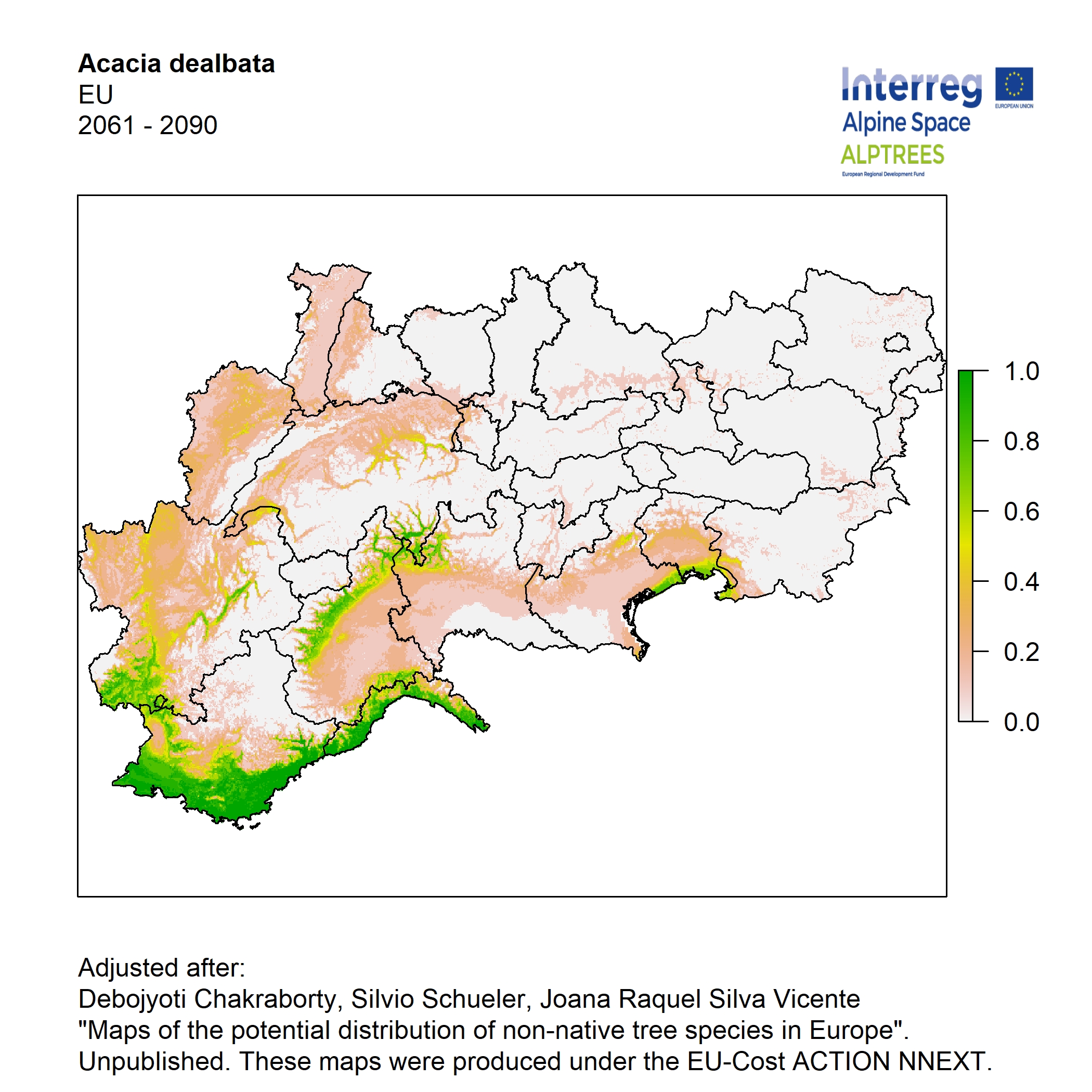
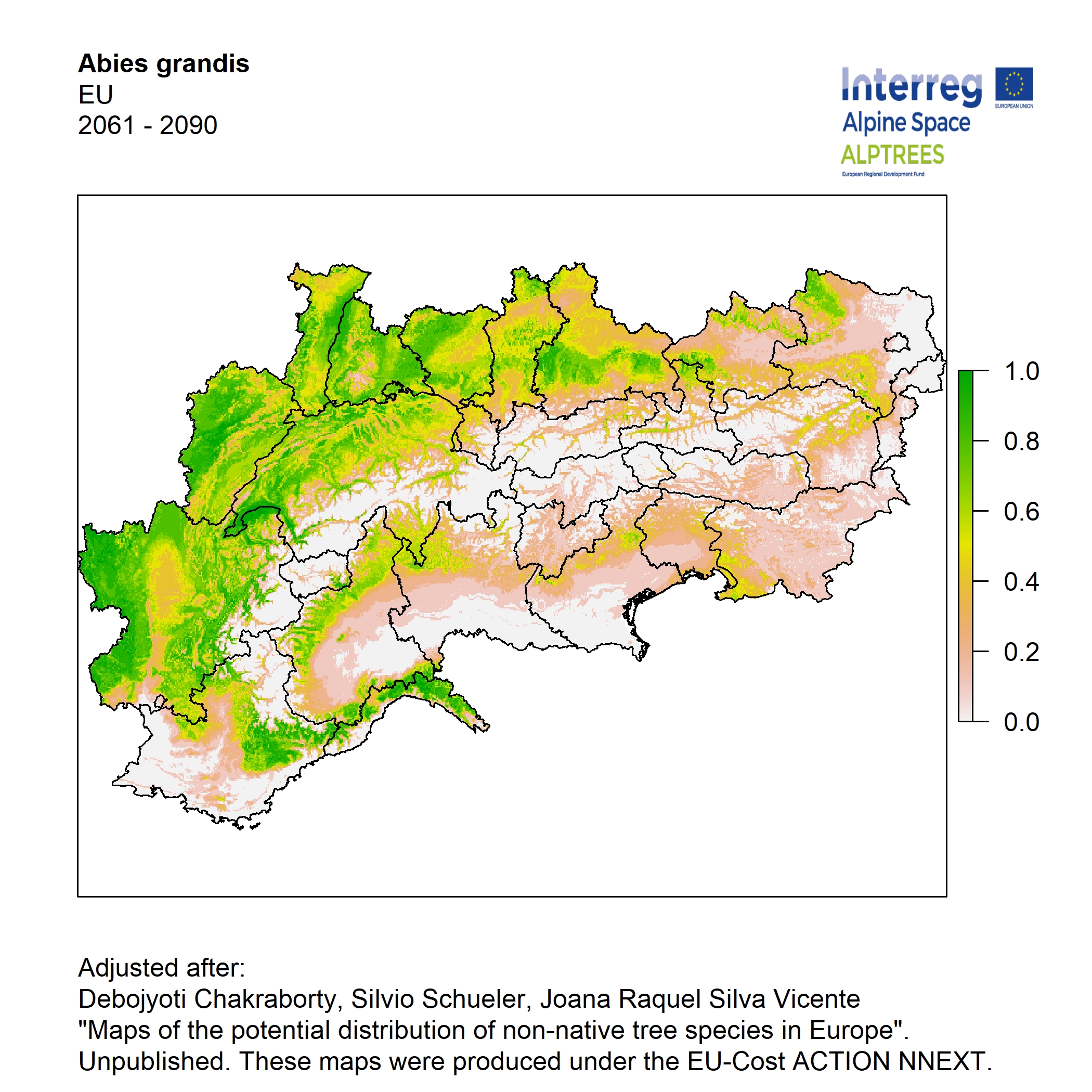
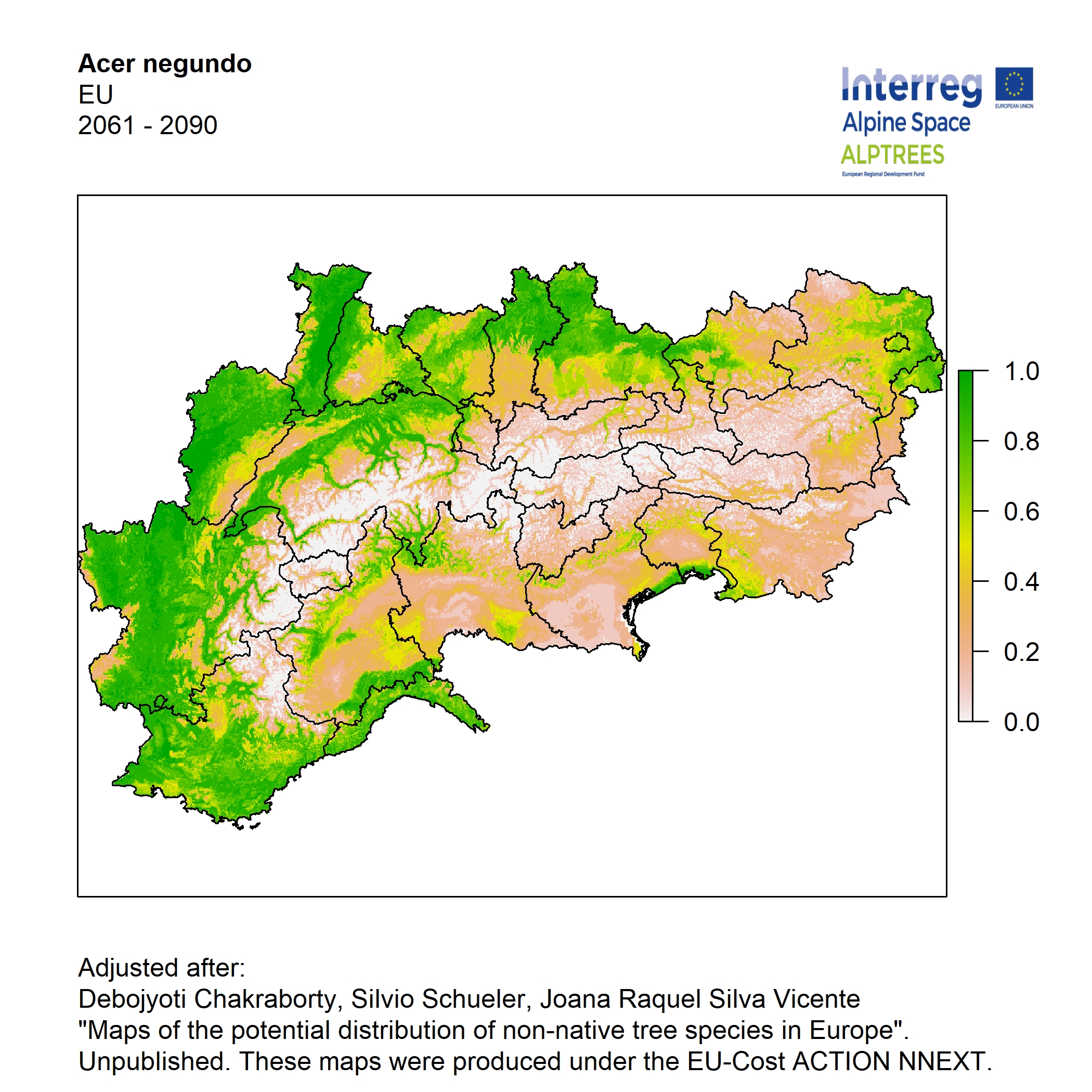
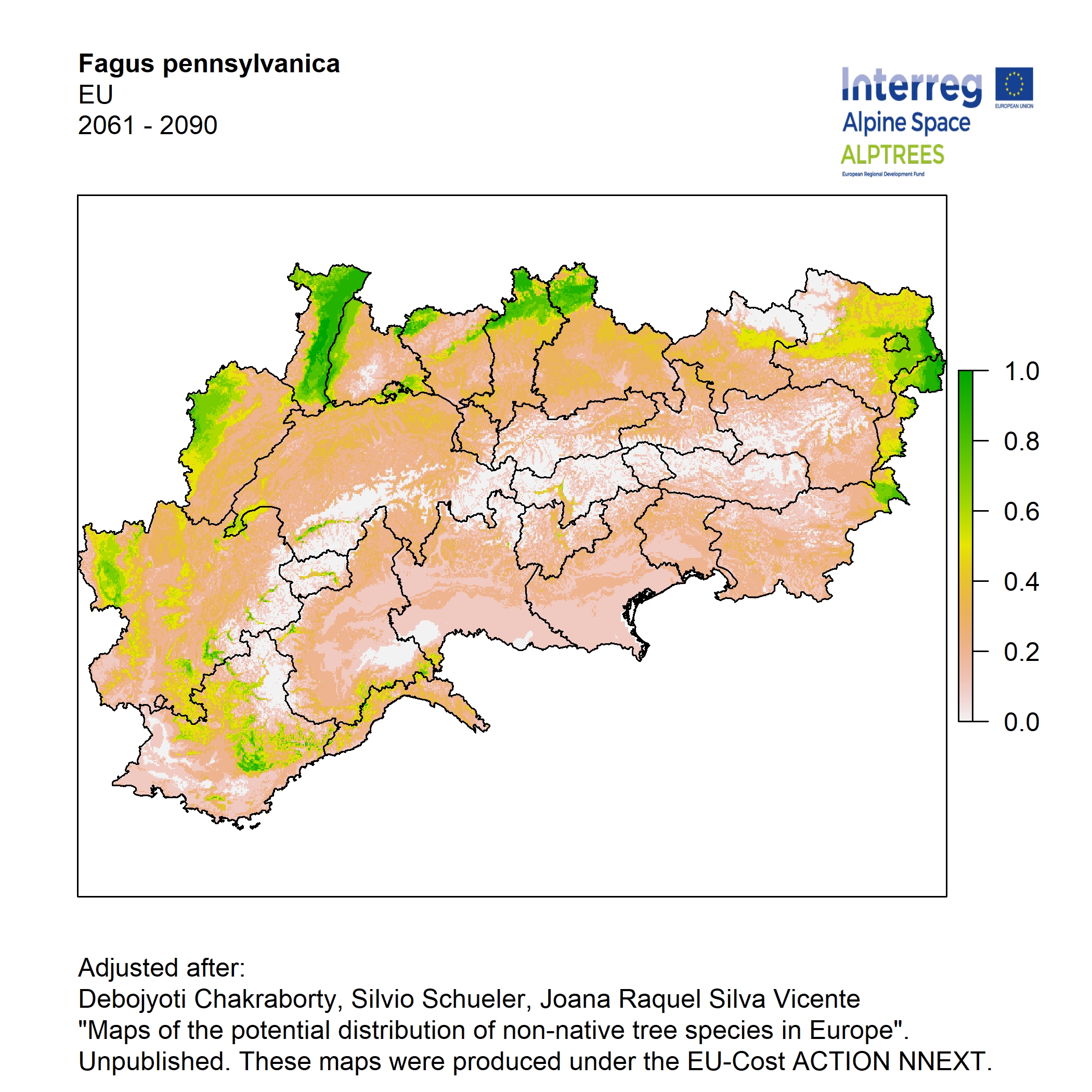
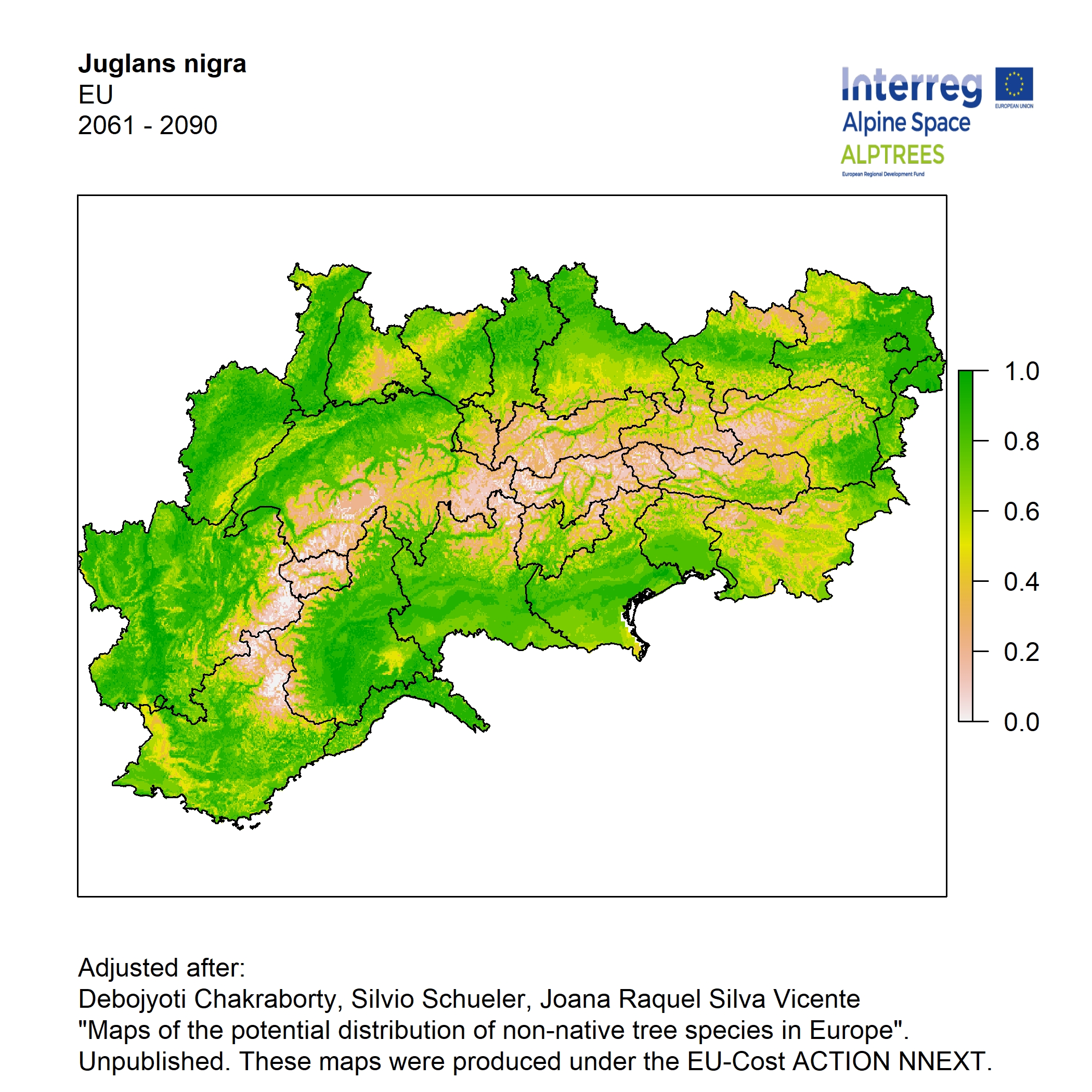
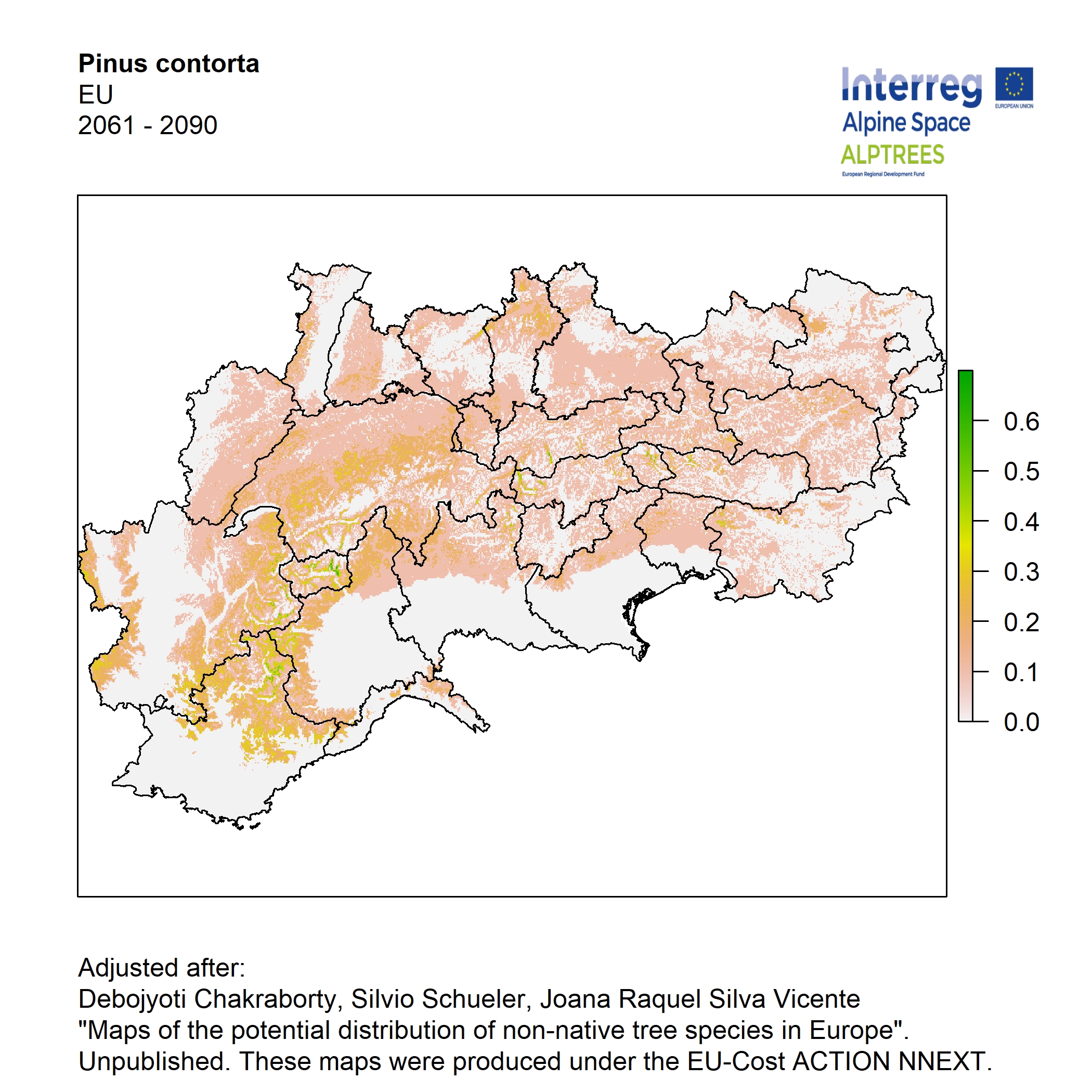
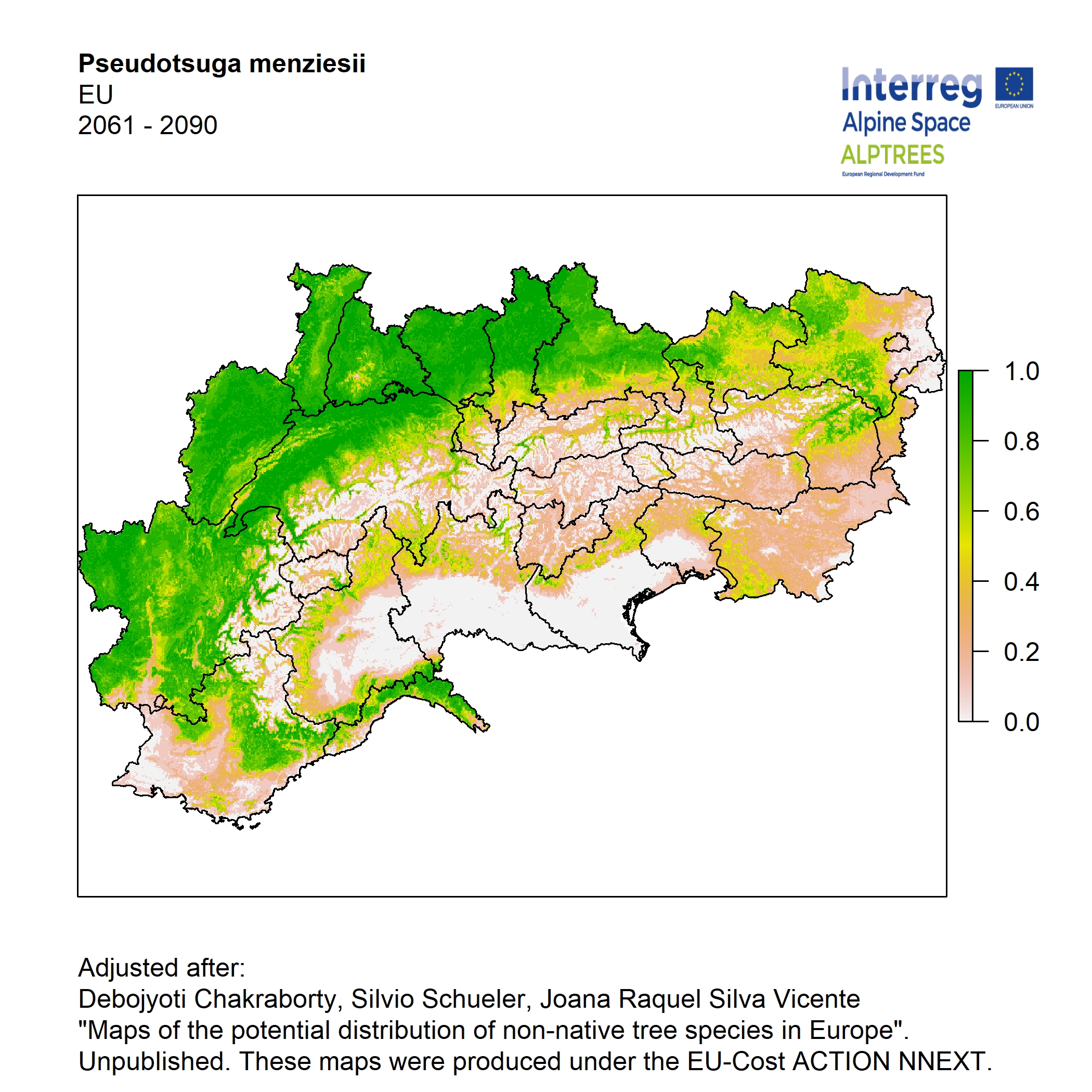
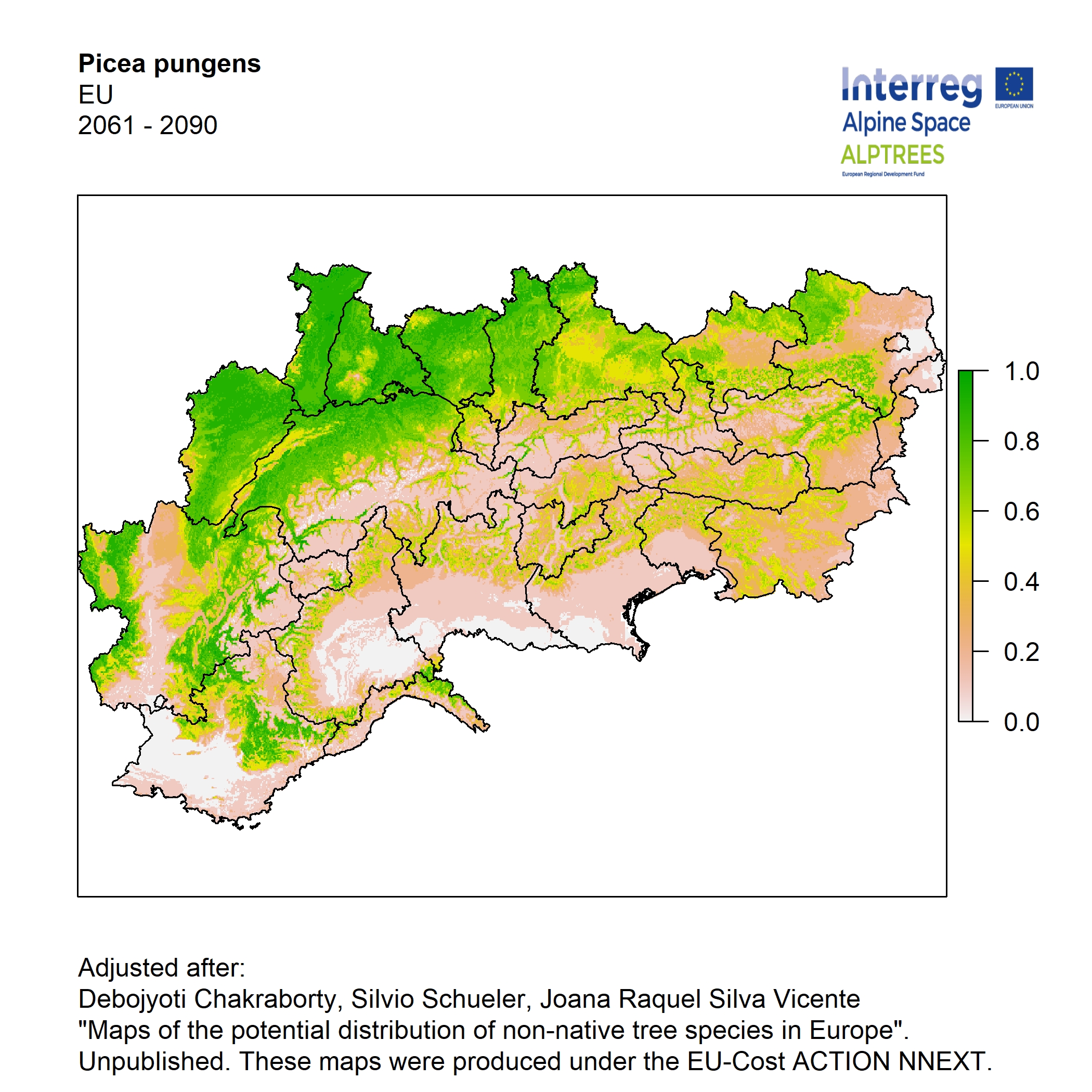
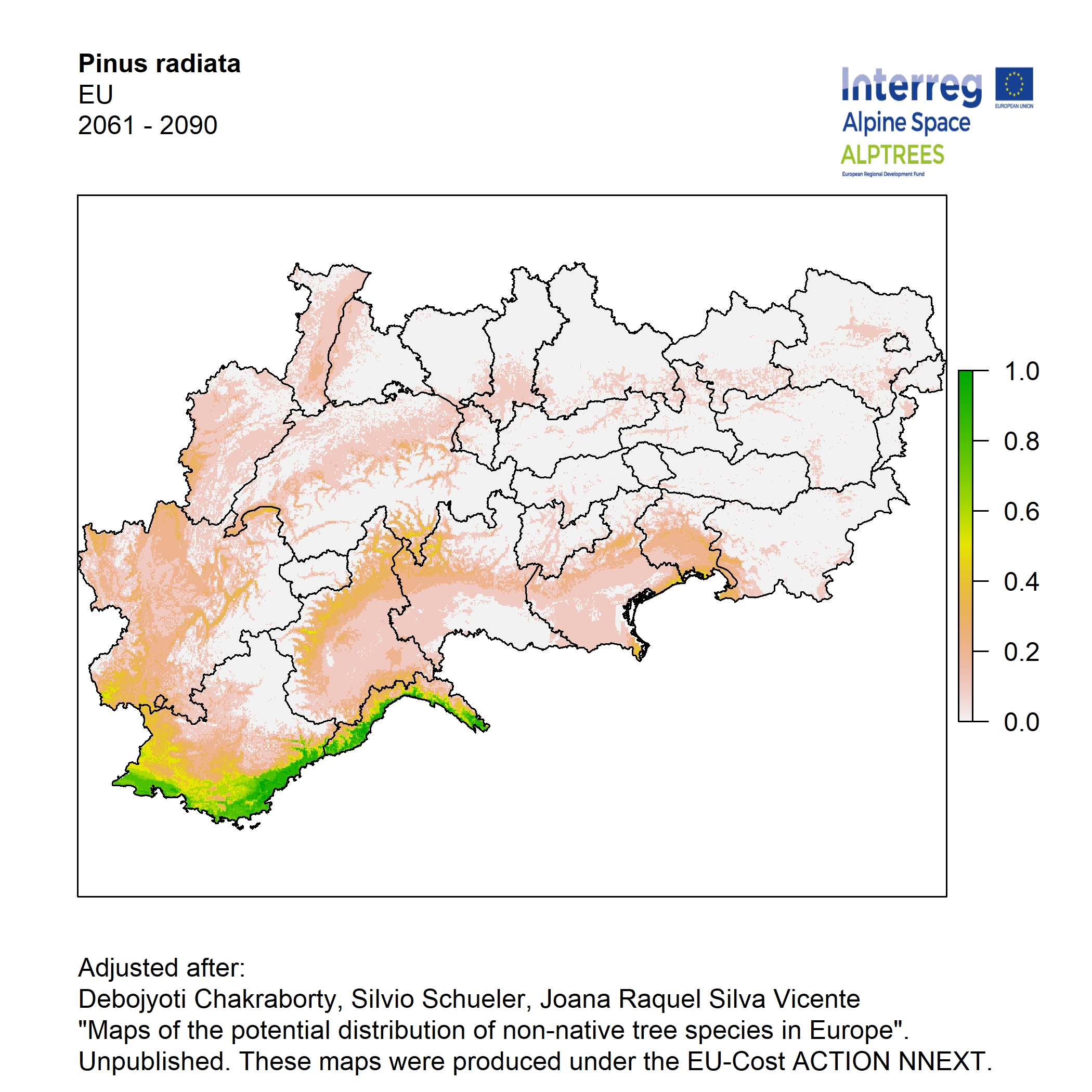
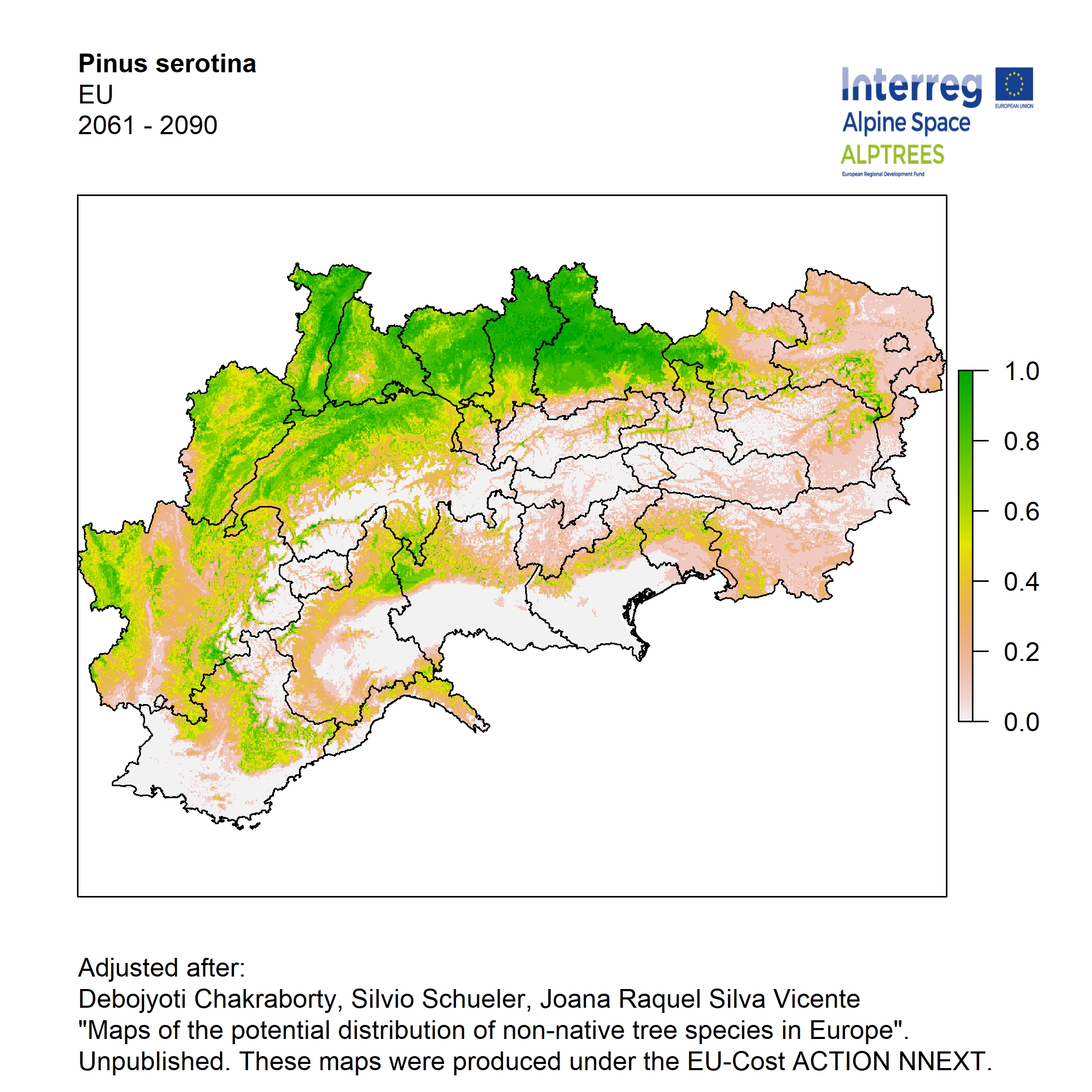
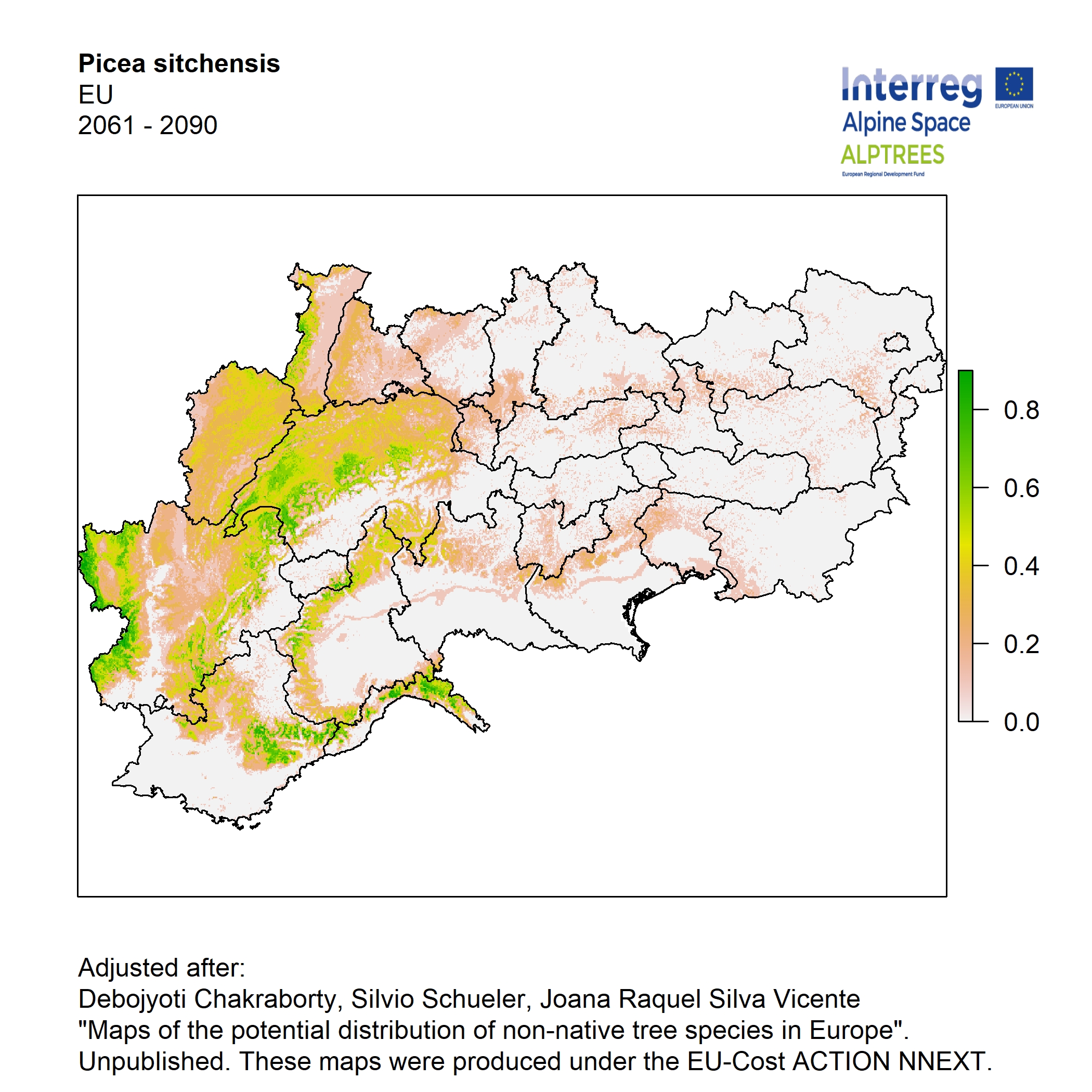
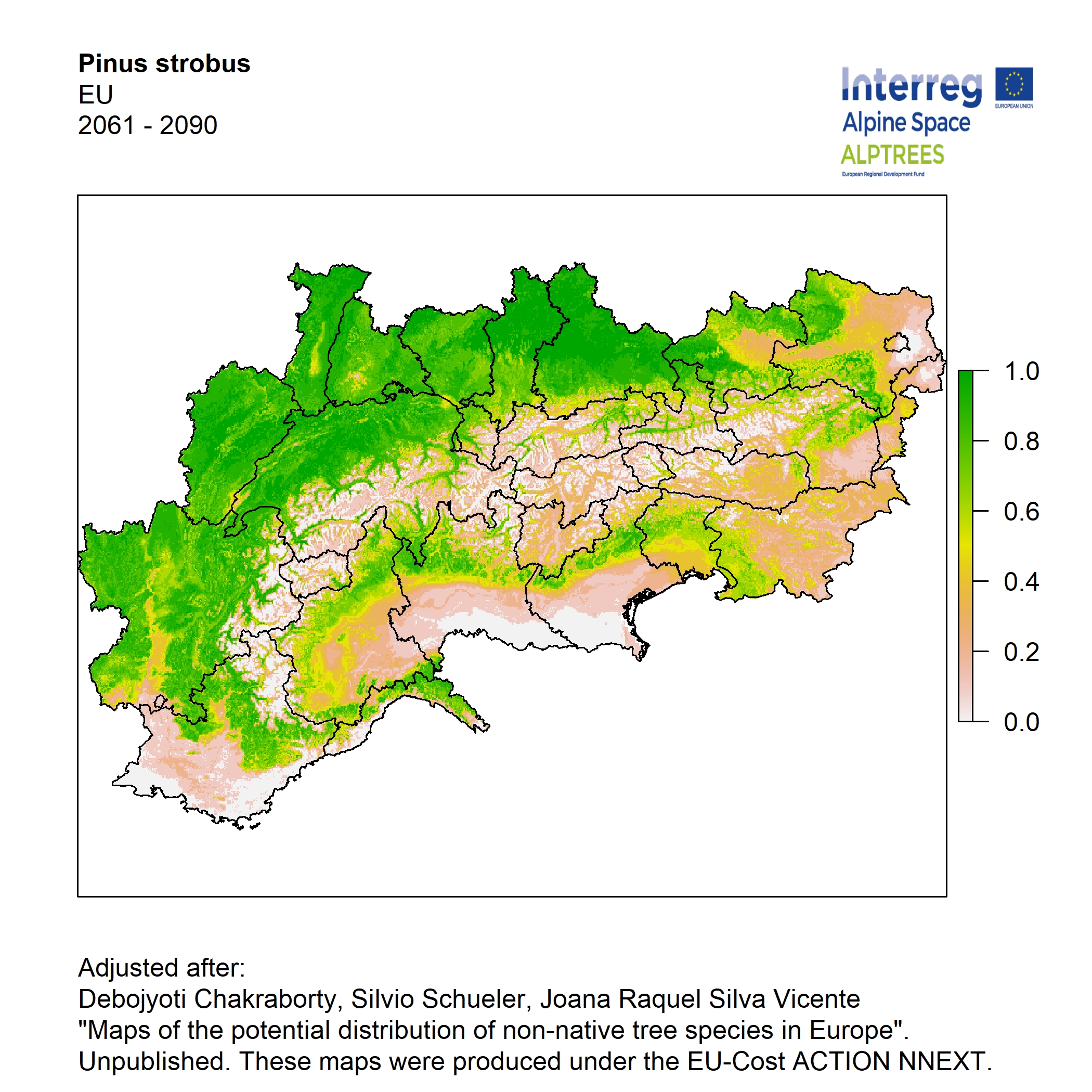
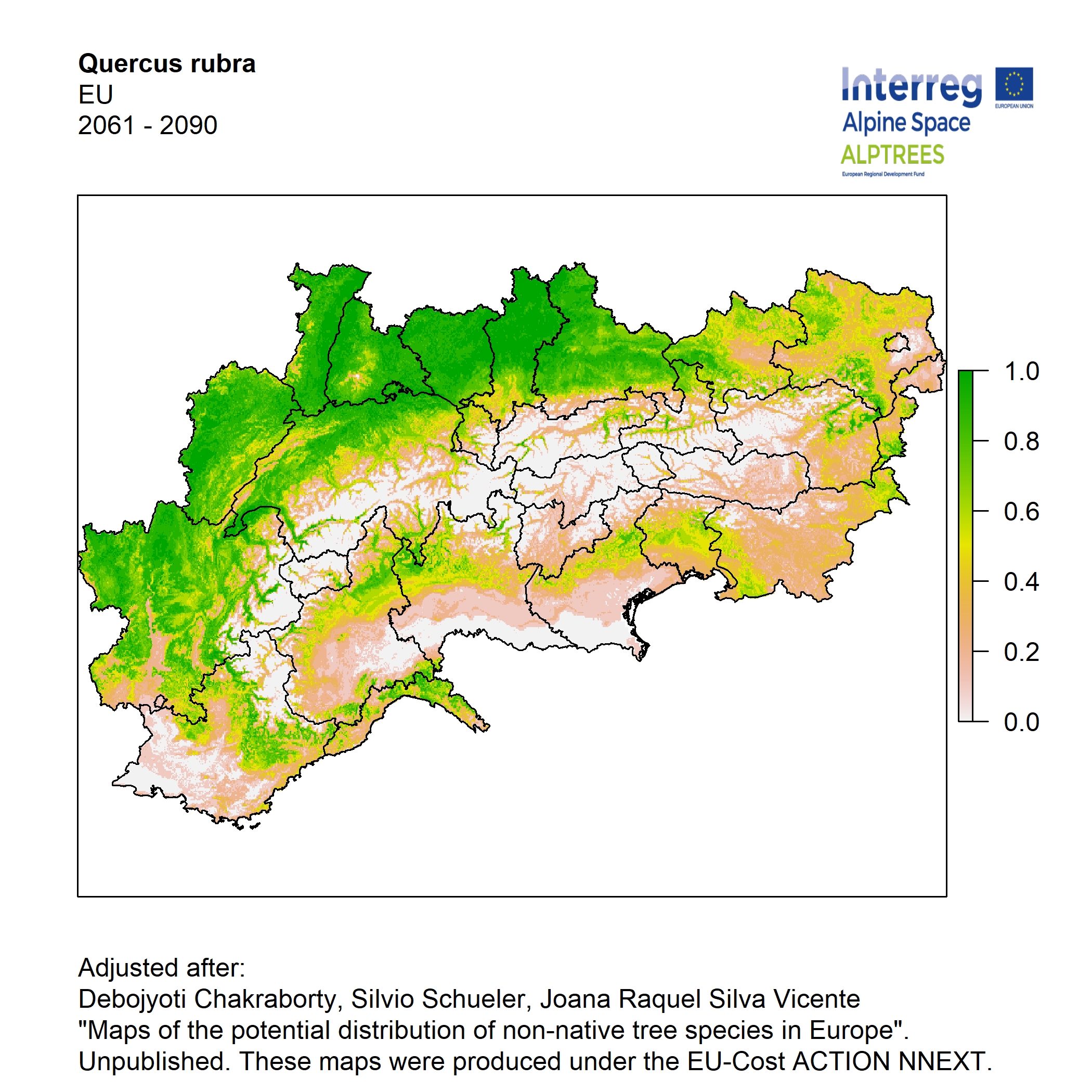
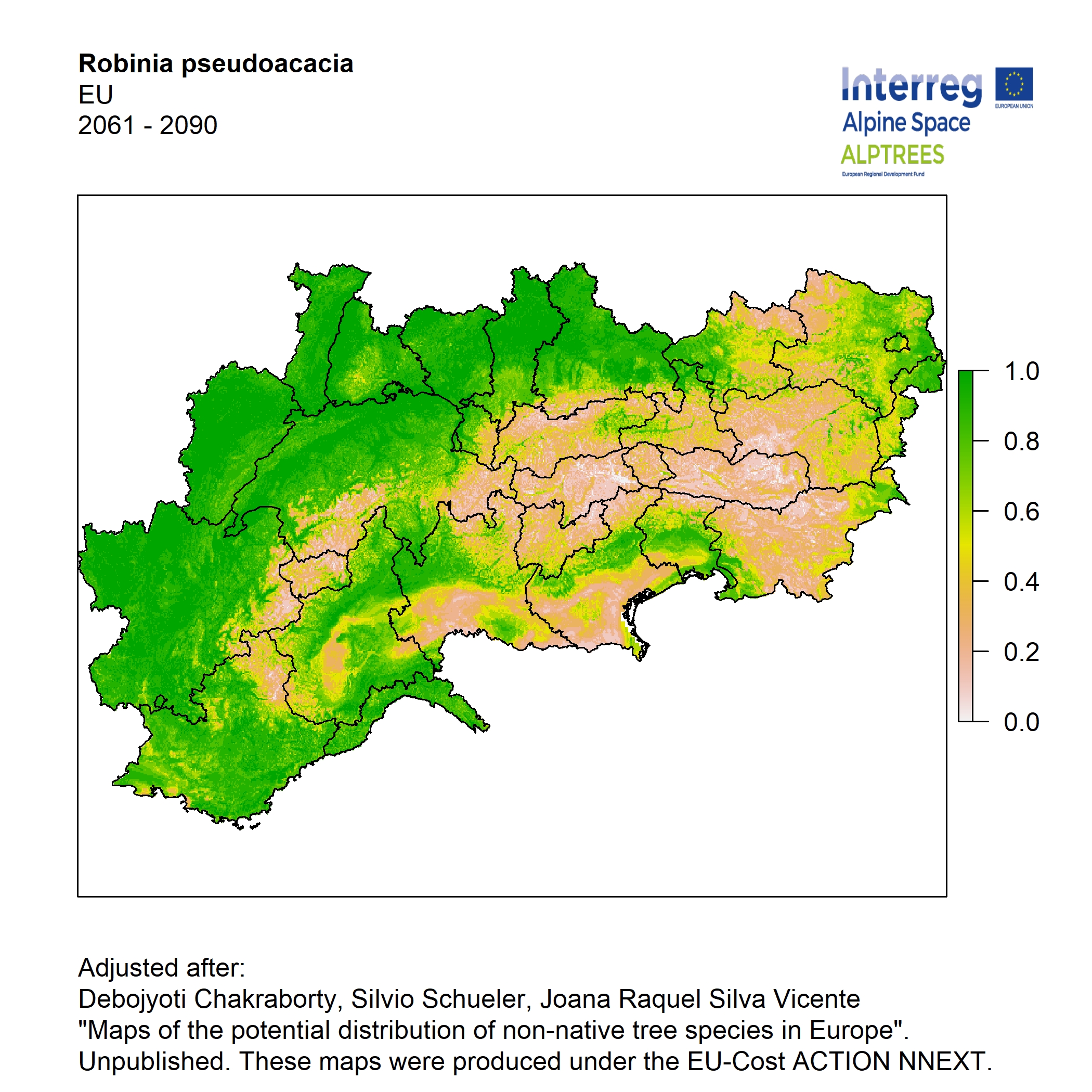
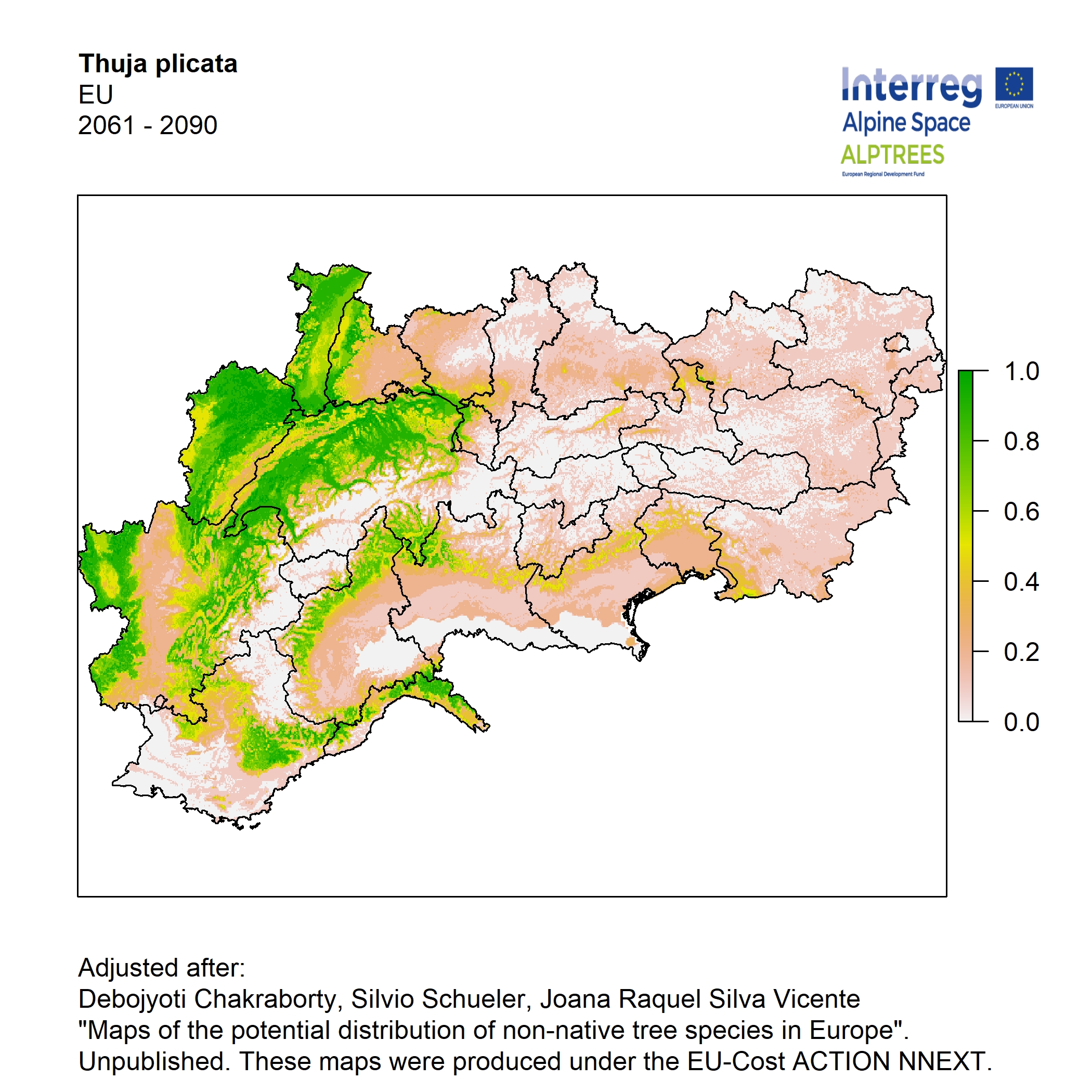
Potential distribution of the NNTs modeled with an ensemble distribution modeling approach, implemented through the R package, Biomod2 (Thuiller et al. 2016). Biomod2 offers a computational platform for multi-method modeling that generates models of species’ potential distribution for each species. The model algorithms include GLM (Generalized Linear Models), GAM (Generalized Additive Models), GBM (Generalized Boosted regression Models), CTA (Classification Tree Analysis), ANN (Artificial Neural Networks), SRE (Surface Range Envelop or BIOCLIM), FDA (Flexible Discriminant Analysis), MARS (Multivariate Adaptive Regression Spline), RF (Random Forest for classification and regression), and MAXENT. Tsuruoka.
For calibrating the models occurrence data from both native and introduced range of the species was used. This approach ensures that a wide range of potential growing considerations of the species is taken into account. Climate data was obtained from (ECLIPS 2.0) Chakraborty et al 2021.
Each model algorithm predicted the probability of the potential distribution for each species. Such probabilities predicted from the individual models were ensembled into a consensus model by combining the median probability over the selected models with True Skill Statistics threshold (TSS > 0.7) (Allouche et al. 2006; Coetzee et al. 2009). The ensemble models were predicted for past and current time frames 1961-1990,1991-2010, and future periods such as 2020-2040,2060-2080,2080-2100 for two climate change scenarios RCP 4.5 and 8.5.